Aptamers have functional similarities with naturally occurring ribozymes and riboswitches, and they offer several advantages over antibodies. Additionally, they can be miniaturized to around 40 bases—around 15 kilodaltons in molecular weight—making them significantly smaller than antibodies.
Breadcrumb navigation
Activities
Nucleic acid aptamers are single-stranded DNA or RNA molecules that can fold into complex three-dimensional shapes to bind specifically to target molecules. These aptamers are created using an evolutionary engineering technique.
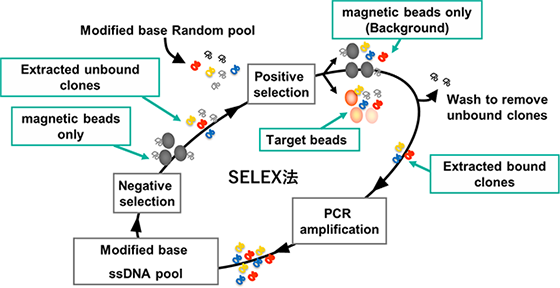
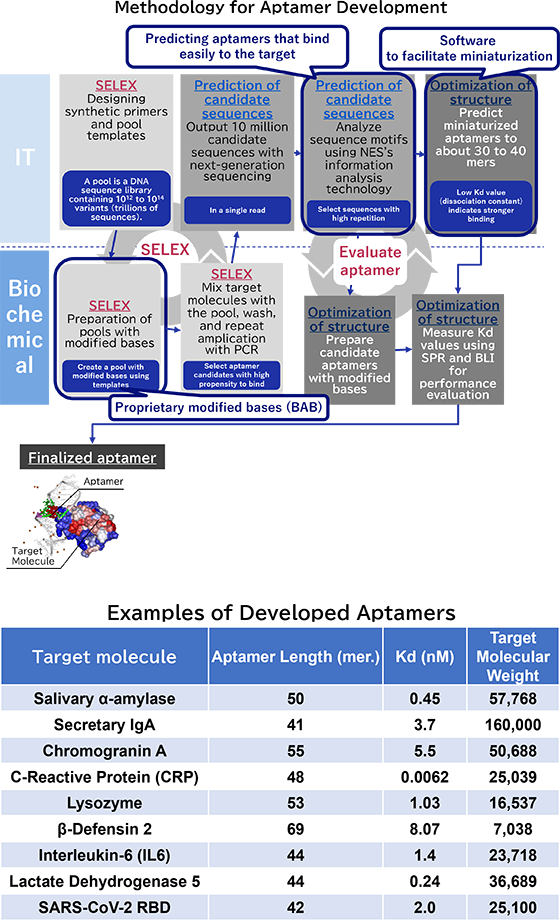
Aptamers are created using an evolutionary engineering technique known as SELEX (Systematic Evolution of Ligands by EXponential enrichment). The process begins with a pool of single-stranded DNA containing a diverse array of sequences. This DNA pool is mixed with beads that have the target molecule immobilized on them. Non-binding DNA is washed away, and the DNA strands that remain bound to the target (known as clones) are isolated. These clones are then amplified using PCR to create the pool for the next round.
This cycle is repeated multiple times, with adjustments made to the number of washes and the amount of target molecule used, thereby progressively enriching the pool for clones that bind strongly to the target.
Development of Base-Appended Base (BAB)-Modified DNA Suitable for Aptamers
Typically aptamers are created from natural DNA or RNA (natural nucleic acids). However, to produce aptamers that bind strongly to specific targets, it is sometimes necessary to use chemically synthesized bases (modified bases) that have other molecules, like amino acids, attached to them.
In collaboration with Dr. Kuwabara of Gunma University (currently at Nihon University), and with support from the Japan Science and Technology Agency (JST) through its A-STEP program, we successfully synthesized a new base-appended base-modified DNA (BAB-DNA) by attaching modified bases to natural DNA. This innovation has been patented (Patent No. 6763551, among others).
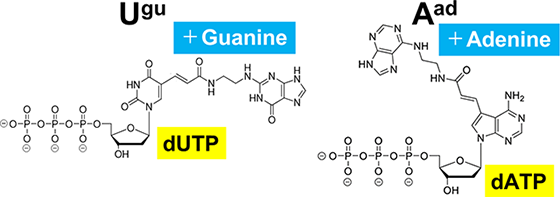
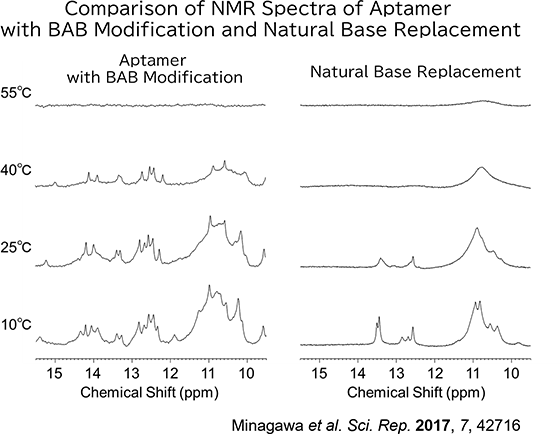
It is known that nucleic acid aptamers containing base-appended bases (BAB) can adopt more complex structures compared to those with the same sequence where the BAB segments have been subsituted with natural bases.
When the BAB segment are replaced with natural bases, the resulting aptamers lose their ability to bind to the target entirely. This suggests that the complex structure enabled by the BAB modification is crucial for the aptamer's binding to its target.
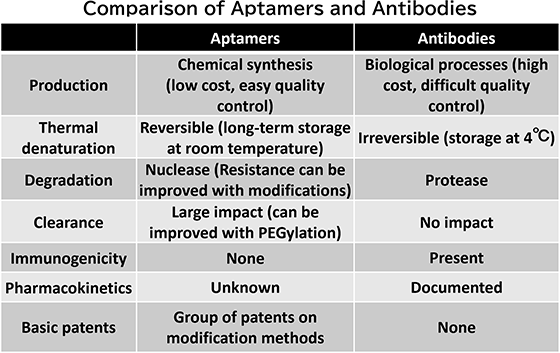
Characteristics of Aptamer Development at NES and Previously Developed Aptamers
We can efficiently develop aptamers from DNA pools containing 1012 to 1014 variants through a combination of biochemical experiments and advanced data analysis techniques. Our key strengths include:
- Employing proprietary modified bases in our biochemical experiments specifically designed for aptamer development
- Utilizing advanced analytical methods to explore and optimize extensive candidate aptamer sequences
Development of Detection Technology Using Aptamers
We are also researching technology to detect target molecules using aptamers.
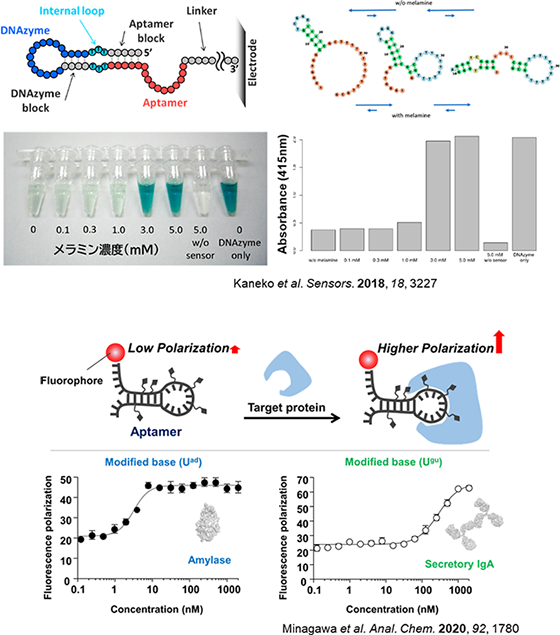
By combing aptamers with DNAzymes, which function like enzymes, we have developed a system where the DNAzyme activates and produces a color change when the target molecule binds to the aptamer. Additionally, leveraging the small size of aptamers, we have developed detection methods using fluorescence polarization to identify target molecules.